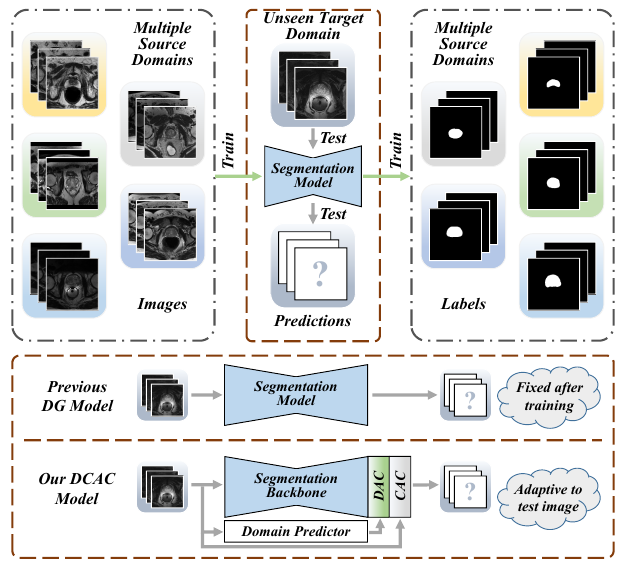
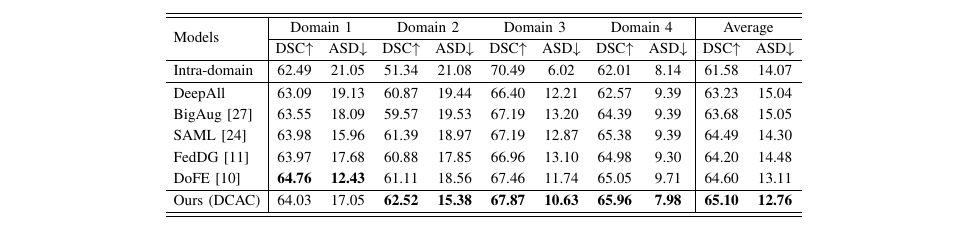
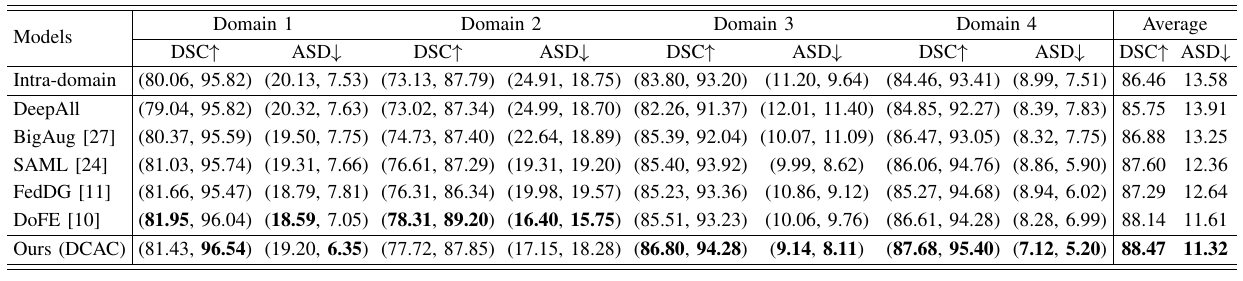
Figure 1: Example of Multi-source Domain Generalization in Medical Image Segmentation.
Abstract
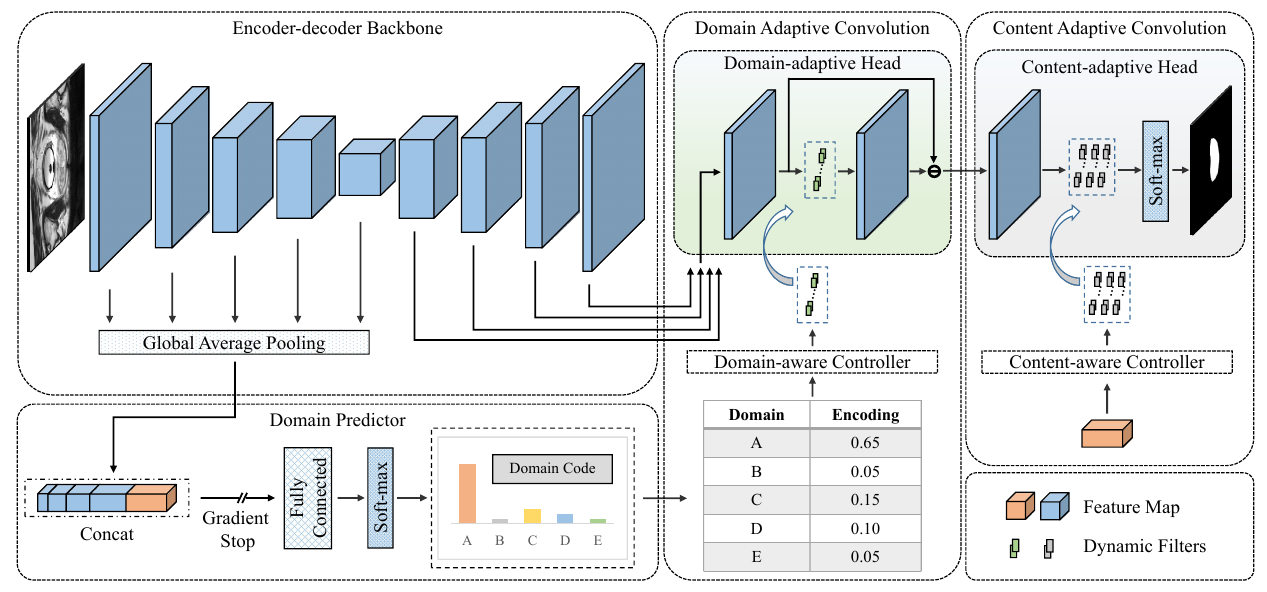
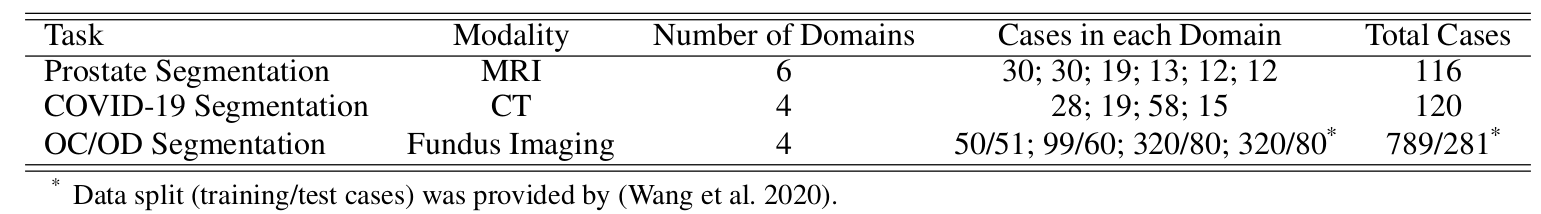
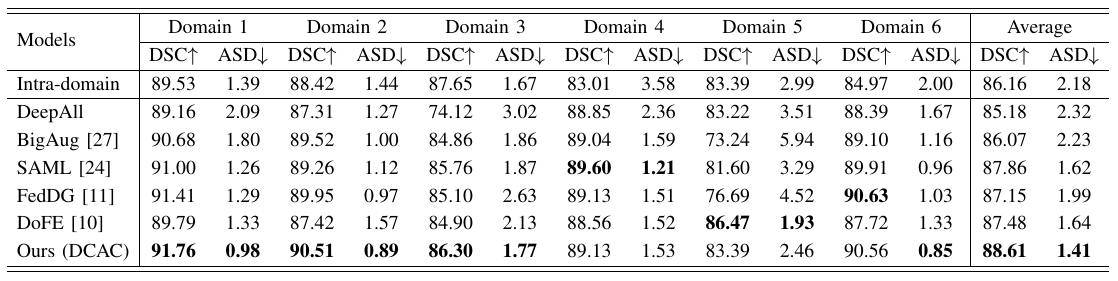
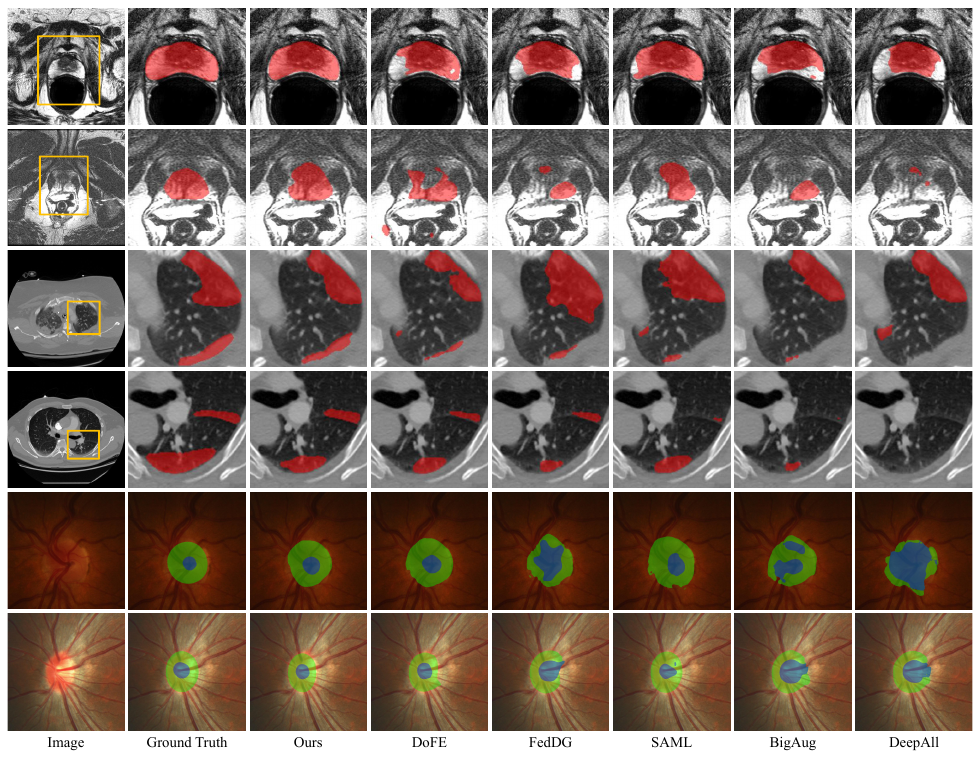
The domain gap caused mainly by variable medical image quality renders a major obstacle on the path between training a segmentation model in the lab and applying the trained model to unseen clinical data. To address this issue, domain generalization methods have been proposed, which however usually use static convolutions and are less flexible. In this paper, we propose a multi-source domain generalization model, namely domain and content adaptive convolution (DCAC), for medical image segmentation. Specifically, we design the domain adaptive convolution (DAC) module and content adaptive convolution (CAC) module and incorporate both into an encoder-decoder backbone. In the DAC module, a dynamic convolutional head is conditioned on the predicted domain code of the input to make our model adapt to the unseen target domain. In the CAC module, a dynamic convolutional head is conditioned on the global image features to make our model adapt to the test image. We evaluated the DCAC model against the baseline and four state-of-the-art domain generalization methods on the prostate segmentation, COVID-19 lesion segmentation, and optic cup/optic disc segmentation tasks. Our results indicate that the proposed DCAC model outperforms all competing methods on each segmentation task, and also demonstrate the effectiveness of the DAC and CAC modules.